Presentation "COBS: A Compact Bit-Sliced Signature Index" at SPIRE 2019 (Best Paper Award)
Posted on 2019-10-08 15:30 by Timo Bingmann at Permlink with 0 Comments. Tags: #talk #university
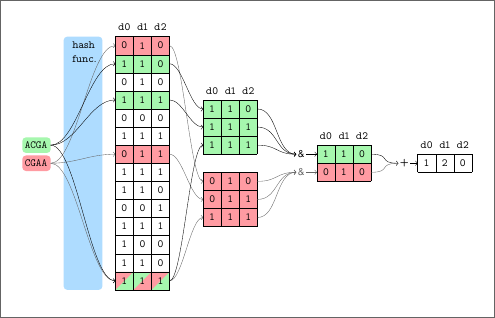
Today, I presented our paper "COBS: A Compact Bit-Sliced Signature Index" at the 26th International Symposium on String Processing and Information Retrieval (SPIRE) 2019 in Segovia, Spain. The full paper is available from this webpage:
paper-COBS-A-Compact-Bit-Sliced-Signature-Index.pdf ,
from the Springer proceedings in LNCS (same content, somewhat differently typeset), and also from arXiv:1905.09624.
I am honored that our paper won the best paper award at SPIRE'19: bestPaperAward.pdf
The slides of my presentation at the SPIRE conference are available here: slides-20191008-cobs-spire.pdf .
The source code and more documentation about COBS can be found on GitHub:
https://github.com/bingmann/cobs.
Update 2019-11-06: COBS now has a Python interface and can be downloaded from PyPI.Abstract
We present COBS, a COmpact Bit-sliced Signature index, which is a cross-over between an inverted index and Bloom filters. Our target application is to index k-mers of DNA samples or q-grams from text documents and process approximate pattern matching queries on the corpus with a user-chosen coverage threshold. Query results may contain a number of false positives which decreases exponentially with the query length. We compare COBS to seven other index software packages on 100000 microbial DNA samples. COBS' compact but simple data structure outperforms the other indexes in construction time and query performance with Mantis by Pandey et al. in second place. However, unlike Mantis and other previous work, COBS does not need the complete index in RAM and is thus designed to scale to larger document sets.